Chromaseq uses the name of each chromatogram file to determine, directly or indirectly, the name to be given to each sequence and information about primers. You will need to define a naming rule that Chromaseq canuse to extract from the name of each chromatogram that chromatogram's sample code and primer name. Currently, the following styles of file names are supported:
- The file name contains embedded within it a sample code and the primer name, and those pieces can be found and then used to look up (via separate text files or databases) information about the names to call sequences and information about the primers, such as their sequences and the gene fragment to which they correspond. [Note: database support not yet available; if it is important to you, please contact
 ]
]
The file names might look like this:
A01_A01DNA1120_B1490_411940.ab1
In this file, the sample code is "1120", and the primer name is "B1490". A separate sample codes and names file will have a list of sample codes and sequence names, and will allow Chromaseq to look up the sequence name to use for sample code "1120"; there will similarly be a primer information file. However, Chromaseq will first have to be able to find the sample code and the primer name within the chromatogram file's name, and for this you need to give Chromaseq a file naming rule that will tell it how to find these elements.
In the future, the following style of file names may be supported, but it is not currently supported:
- The file names contain only a single code (e.g.," B03"), which is either the cell in a spreadsheet or spreadsheets or database that contains the sample code or primer name, more details about which can be looked up in additional files or database tables.
If this style of file names, or other ones, are the way you link information about the sequence names and primer information to your chromatograms, and it would be important to you for Chromaseq to support these, then please contact ![]() .
.
Defining naming rules
To select a rule that specifies how the file names are structured, you will use the following dialog box, which will appear when you choose one of the menu items to process chromatograms:
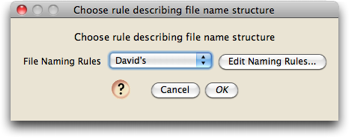
When you first install Chromaseq, the only option will be "default", and you will first need to define the naming rule that works for your files. To do this, touch on the "Edit Naming Rules" button in the above dialog, and you will be presented with a list of available naming rules (when you first install Chromaseq, that list will likely be empty):
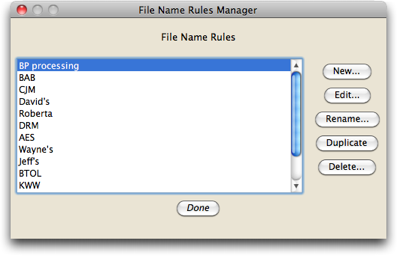
To create a new naming rule, touch on the "New" button. To edit, rename, duplicate, or delete a rule, select it first from the list, then touch on the appropriate button.
If you make a new rule called "myRule", this is the dialog box you will get to edit the naming rule:
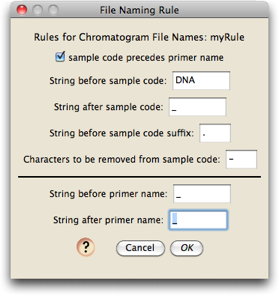
The naming rule shown in the above dialog will work on the sequences mentioned above, whose names look like this:
C11_C11DNA1780_D3aR_583152.ab1
C11_C11DNA1780_NLF184_582960.ab1
E09_E09DNA0252_D3aR_887908.ab1
E09_E09DNA0252_NLF184_887812.ab1
For example, in the first file, "1780", the sample code, has as a string of character before it "DNA", and has the string "_" immediately following it. The primer "D3aR" has an underscore before it, and after it.
Variants
If you wish some characters to be automatically removed from the sample code, then enter these characters in the "Characters to be removed from sample code" box. For example, in the sequencing facility that DRM uses, hyphens are occasionally (and apparently randomly) inserted in the file name as part of the sample code, and so those need to be automatically stripped out.
In some cases, the sample code name consists of two parts, a main sample code, and a suffix. For example, the following files are chromatograms from four clones (p, q, t, and u) of sample 1794.
C03_C03CL1794.p_B1490_287986.ab1
C03_C03CL1794.p_Pat_288082.ab1
C04_C04CL1794.q_B1490_287994.ab1
C04_C04CL1794.q_Pat_288090.ab1
C05_C05CL1794.t_B1490_288002.ab1
C05_C05CL1794.t_Pat_288098.ab1
C06_C06CL1794.u_B1490_288010.ab1
C06_C06CL1794.u_Pat_288106.ab1
The sample code thus consists of two parts , "1794", and the suffix p, q, t, or u. The string before the sample code suffix is ".". If Mesquite encounters in the sample code a string that matches that designated in "string before sample code suffix", then it will split the sample code name in two, and consider the main sample code to be the first part.
If you have asked to have the sample code translated into full sample names, then Mesquite will use just the part before the suffix (if there is one) as the code to look up in the sample names file, but it will append the suffix on to the end of the full sample name. For example, for these two files:
C05_C05CL1794.t_B1490_288002.ab1
C05_C05CL1794.t_Pat_288098.ab1
the sequence produced will be a COI sequence (primers Pat and B1490), named "Bembidion integrum IL 1490 t", as "Bembidion integrum IL 1490" is the sample name listed in the sample names file.
If the sample code is at the very start of the file name, set the string before sample code to be blank.
Note: only one naming rule is currently allowed for all files processed at one time.
Another example naming rule
In the following file names, the sample code is at the very start of the file name, and is followed by a "#":
d124#050302-12261.ab1
d096#041117-12261.ab1
d124#050302-13398.ab1
d096#041117-13398.ab1
The primer names in these are the "12261" and the "13398"; the primer name follows a "-" and precedes a ".". Thus, the naming rule for files with these sorts of names are:
- sample code precedes the primer name
- String before sample code is "" (thus, it is at the very start of the file name). Make sure there is no space in the field; there should be absolutely nothing in there.
- String after sample code is "#"
- String before primer name is "-".
- String after primer name is "."